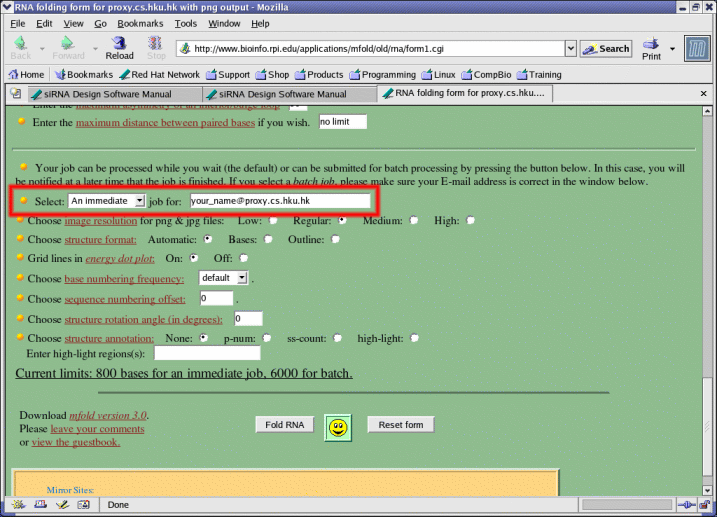
- At least one software must be selected.
- Some software invokes BLAST by default and requires a longer processing time. A longer timeout has been set by default for these software but users can also change the individual timeout for each software if needed.
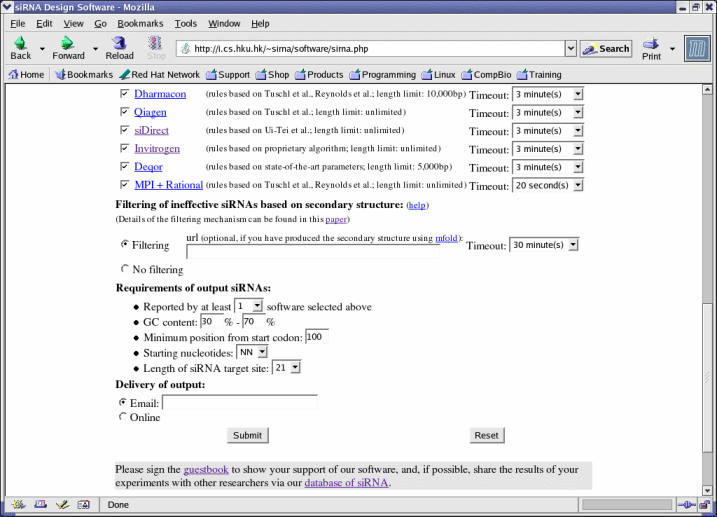
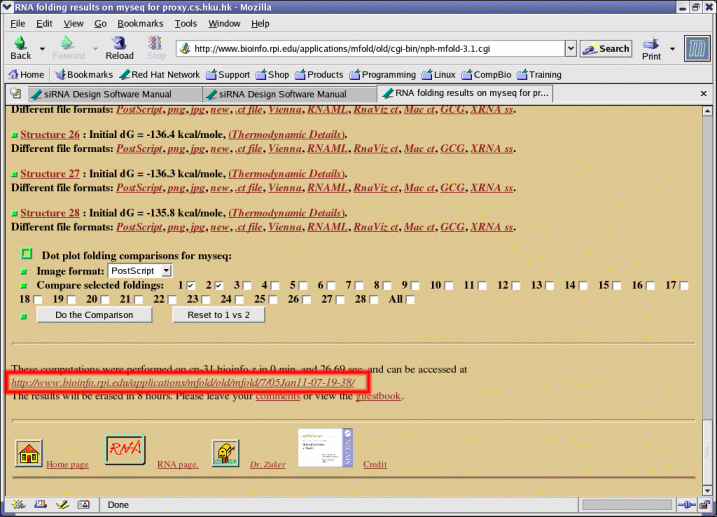
- To use mfold, users can also choose to provide an optional url containing the mfold results (see Appendix II). If no url is present, mfold will be invoked automatically before filtering and a longer time (around 30 minutes) is needed.
- Users can also specify a longer timeout if the default timeout is not long enough for their sequence.
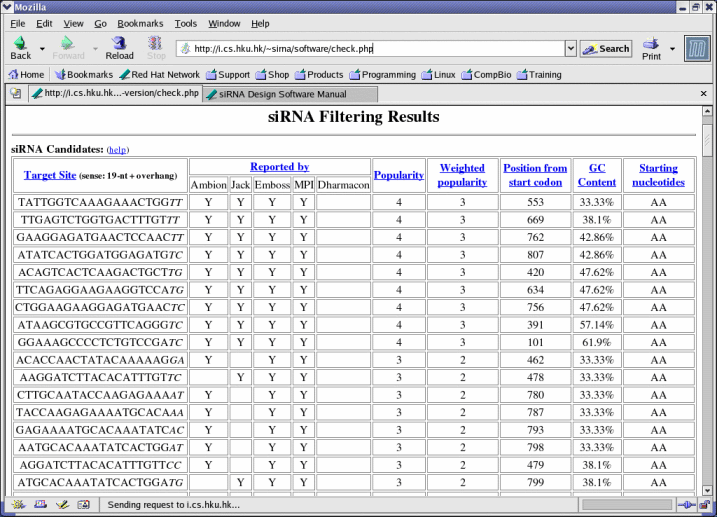
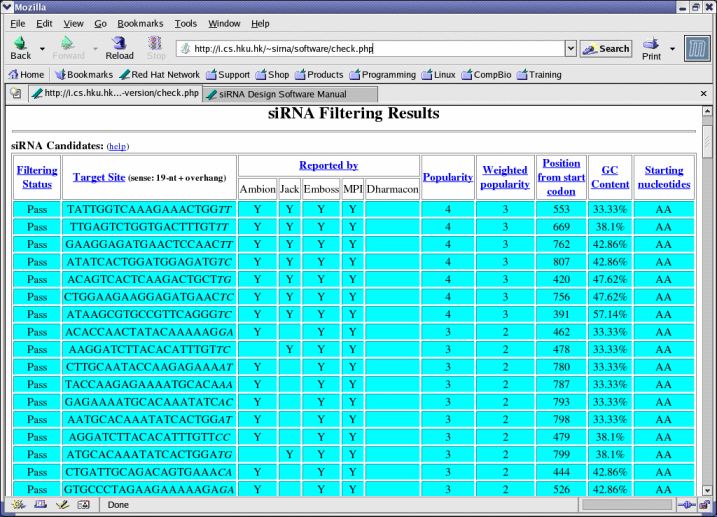
- The number of software selected here should not exceed the number of software selected in Software. This number is also called the popularity of an siRNA candidate.
- GC content should be within the range 0%-100%.
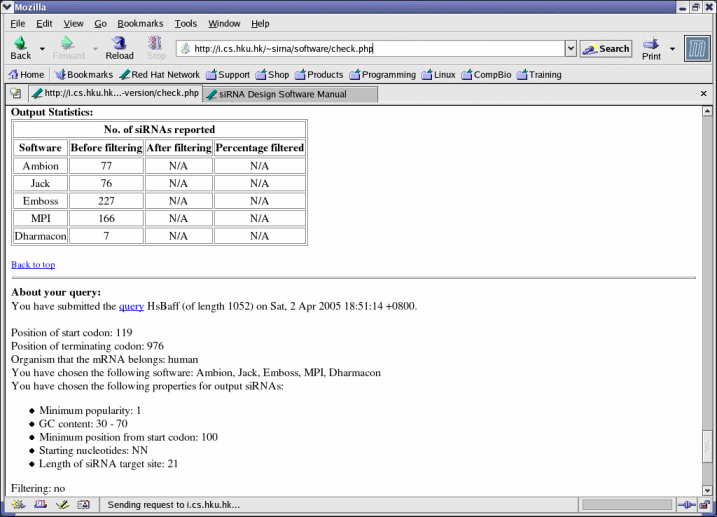
- The minimum position from start codon should be within the range (-start codon + 1) to (terminating codon - start codon). For example, if the start codon is at 76 and the terminating codon is at 963, then the minimum position from the start codon should be within the range -75 to 887.
- Starting nucleotides is the dinucleotide sequence just before the target site (such as AA, NA, where "N" refers to any nucleotide). For example, if the target sequence starts from position 76, the two nucleotides in positions 74 and 75 are its starting nucleotides.
- The default length of the siRNA target site is 21. Other lengths are not supported at the current moment.
- To receive results via email, a valid email address should be provided. The email address is kept confidential and will be discarded once the result is sent to the address. A link to the result page will be provide in the email. The results will be erased after 48 hours.
- Results can also be displayed directly after processing if "Online" is selected.